“You’re doing something really important with people who are important to you,” Paul Worsley remarks when asked about having his younger brothers Caleb and Adam as lab mates. The trio are undergraduate students working in the lab of Santimukul Santra, Ph.D., at Pittsburg State University in Pittsburg, Kansas.
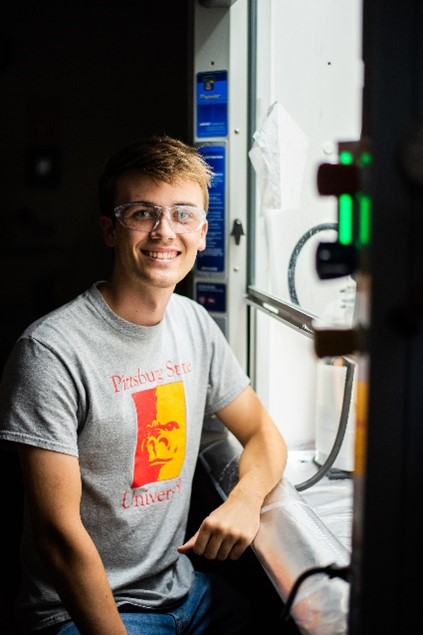
All three brothers are part of the Kansas IDeA Networks of Biomedical Research Excellence (K-INBRE). Paul is currently a junior majoring in biology and history. He plans to go to medical school when he graduates, but his time in the lab has given him a love for research—and has even led him to toy with the idea of going to graduate school instead. His twin brothers Caleb and Adam are only freshmen, but they both think they want to pursue scientific research when they graduate.
When Paul was a sophomore, he applied for a K-INBRE research spot in Dr. Santra’s lab and was immediately accepted. He quickly realized that organic chemistry in the lab was much different—and more exciting—than anything he’d seen in the classroom. “I like organic synthesis because it really tests your knowledge,” he says. “Answering exam questions is way different than actually doing it in a lab.” Despite the challenges that came with research, Paul was clearly doing great work because one day Dr. Santra joked, “Hey, you got any brothers?” Paul responded, “Actually, yes.”
Continue reading “Three Brothers Are Making Research a Family Affair”


 Dr. Idhaliz Flores-Caldera.
Dr. Idhaliz Flores-Caldera.