Hunting for the cause of a disease can be like tracing a river back to its many sources. Myriad factors, large and small, may contribute to a condition. One approach to the search focuses on the massive amounts of genomic and other biological data that scientists are gathering in the course of their studies. To examine this data and look for meaningful patterns and other clues, scientists turn to bioinformatics, a field focused on the development of analytical methods and software tools.
Here are a few examples of how National Institutes of Health-funded scientists are using bioinformatics to dig deeply into data and learn more about the development of diseases, including Huntington’s, preeclampsia and asthma.
Huntington’s Disease

The cause of Huntington’s disease, a degenerative neurological disorder with no known cure, may appear simple. It begins with a change in a single gene that alters the shape and functioning of the huntingtin protein. But this protein, whether in its normal or altered form, does not act alone. It interacts with other proteins, which in turn interact with others.
A research team led by Robert Hughes of the Buck Institute for Research on Aging set out to understand how this ripple effect contributes to the breakdown in normal cellular function associated with Huntington’s disease. The scientists used experimental and computational approaches to map a network of 2,141 proteins that interact with the huntingtin protein either directly or through one other protein. They found that many of these proteins were involved in cell movement and intercellular communication. Understanding how the huntingtin protein leads to mistakes in these cellular processes could help scientists pursue new approaches to developing treatments.
Preeclampsia
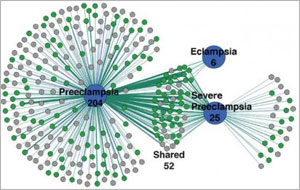
While some diseases, such as Huntington’s, can be traced back to a single gene, others result from many. For example, hundreds of genes contribute to preeclampsia, a potentially life-threatening condition associated with high blood pressure during pregnancy. Using bioinformatics, scientists have identified unique groupings of these genes underlying different forms of the disease.
A research team led by James Padbury of the Women and Infants Hospital of Rhode Island used advanced data-mining techniques to sort through 28,000 records of preeclampsia research in PubMed, an NIH database of scientific articles. After finding more than 2,000 studies on preeclampsia genetics, the team narrowed its search to 729 studies documenting 535 genes that have clear associations with preeclampsia. Using cluster analysis, a tool that reveals patterns in data, the scientists discovered that various forms of preeclampsia, which differ in timing and severity, are associated with distinct groupings of maternal and fetal genes.
The scientists now plan to make their data freely available online in a searchable database so that others can further explore the complex genetic causes of preeclampsia.
Asthma
In addition to genes, environmental and lifestyle factors can contribute to certain medical conditions. A person’s medical history, lung function and exposure to animals or secondhand smoke, for instance, may all contribute to symptoms associated with asthma. To offer more individualized asthma treatments, doctors must figure out which variables are contributing factors for which individuals.
Recently, a team of scientists showed that a computational method capable of analyzing more than 100 of these variables for a large number of people can be used to group asthma patients by the possible root causes of their symptoms. In this study, the research team, led by Wei Wu of Carnegie Mellon University and Sally Wenzel of the University of Pittsburgh, used a computer algorithm developed by Wu to identify several patient clusters. These clusters corresponded to known subtypes of asthma, such as those caused by allergies or sinus disease, but also included a possible new subtype of asthma in which patients have near-normal lung function and yet severe symptoms.
The researchers believe their approach could help improve the understanding and treatment not just of asthma but of many other complex conditions such as osteoporosis, kidney disease and autoimmune diseases.


Keep up the good work, I have been exposed to mold and I have asthma it’s something I wish on no one don’t know what to do Drs do not want to deal with it.