Even though scientists have been studying genetics since the mid-19th century, they continue to make new discoveries about genes and how they impact our health on a regular basis. NIGMS researchers study how genes are expressed and regulated, how gene variants with different “spellings” of their genetic code affect health, and much more. Get the drop on DNA and the gist of genes with these fast facts:
Continue reading “Genetics by the Numbers”Category: Genes
What Is Genetics?
Genetics is the study of genes and heredity—how traits are passed from parents to children through DNA. A gene is a segment of DNA that contains instructions for building one or more molecules that help the body work. Researchers estimate that humans have about 20,000 genes, which account for about 1 percent of our DNA. The remainder of the DNA plays a role in regulating genes, and scientists are researching other potential functions.
Continue reading “What Is Genetics?”Propelling Rare Disease Research for More Than 50 Years

The year 2022 marked 50 years since the creation of the NIGMS Human Genetic Cell Repository (HGCR) at the Coriell Institute for Medical Research in Camden, New Jersey. The NIGMS HGCR consists of cell lines and DNA samples with a focus on those from people with rare, heritable diseases. “Many rare diseases now have treatments because of the samples in the NIGMS HGCR,” says Nahid Turan, Ph.D., Coriell’s chief biobanking officer and co-principal investigator of the NIGMS HGCR. She gives the example of a rare disease advocacy group who worked with the NIGMS HGCR to establish a cell line several decades ago. It was used to identify a gene associated with the disease, which aided in the development of five treatments that have received approval from the Food and Drug Administration.
Researchers have also studied NIGMS HGCR’s samples to help advance knowledge of basic biology and genetics, and even to support the development of a vaccine for a deadly virus.
Continue reading “Propelling Rare Disease Research for More Than 50 Years”Advancing American Indian and Alaska Native Health Through Research, Training, and Engagement
American Indian and Alaska Native (AI/AN) populations have long experienced health disparities such as higher rates of diabetes, certain cancers, and mental health conditions than those of other Americans. One contributing factor in these disparities is underrepresentation of AI/AN populations in biomedical science—as study participants, researchers, and health professionals. Unfamiliarity with health care options and opportunities, coupled with a distrust of biomedical research resulting from unethical studies in the past, have exacerbated this underrepresentation.
NIGMS-supported researchers, including Native scientists, are partnering with AI/AN Tribes to help reduce health disparities by conducting research focused on AI/AN health priorities and building infrastructure that supports research in those communities. They’re also preparing Native students to pursue careers in science and medicine. In this post, you’ll meet four scientists advancing AI/AN health.
Continue reading “Advancing American Indian and Alaska Native Health Through Research, Training, and Engagement”Career Conversations: Q&A with Biochemist Alexis Komor

“DNA is an amazingly beautiful molecule, and it’s so important. Each of our cells has only one copy of DNA, and if it gets damaged, that messes up everything else in the cell,” says Alexis Komor, Ph.D., an assistant professor of chemistry and biochemistry at the University of California, San Diego (UCSD). Check out the highlights of our interview with Dr. Komor to learn about her scientific journey, research on DNA, and advice for students.
Q: How did you decide to study chemistry?
A: I really enjoyed math and science in middle and high school. When I applied to college, I knew I wanted to major in science over math because I felt like it was more relevant to what we experience on a day-to-day basis. I ultimately went into chemistry for a silly reason, but looking back now, I’m so very grateful that I did. Chemistry has this nice balance because it allows you to not only understand how things work on a molecular level but also see how those molecular workings relate to everyday phenomena—for example, understanding how DNA damage on a molecular level can lead to negative health outcomes.
Continue reading “Career Conversations: Q&A with Biochemist Alexis Komor”Hunting Disease-Causing Genetic Variants

“In my lab, we’ve been gene hunters—starting with visible phenotypes, or characteristics, and searching for the responsible genes,” says Miriam Meisler, Ph.D., the Myron Levine Distinguished University Professor at the University of Michigan Medical School in Ann Arbor. During her career, Dr. Meisler has identified the functions of multiple genes and has shown how genetic variants, or mutations, can impact human health.
Becoming a Scientist
Dr. Meisler had a strong interest in science as a child, which she credits to “growing up at the time of Sputnik” and receiving encouragement from her father and excellent science teachers in high school and college. However, when she started her undergraduate studies at Antioch College in Yellow Spring, Ohio, she decided to explore the humanities and social sciences. After 2 years of sociology and anthropology classes, she returned to biomedical science and, at a student swap, symbolically traded her dictionary for a slide rule—a mechanical device used to do calculations that was eventually replaced by the electric calculator.
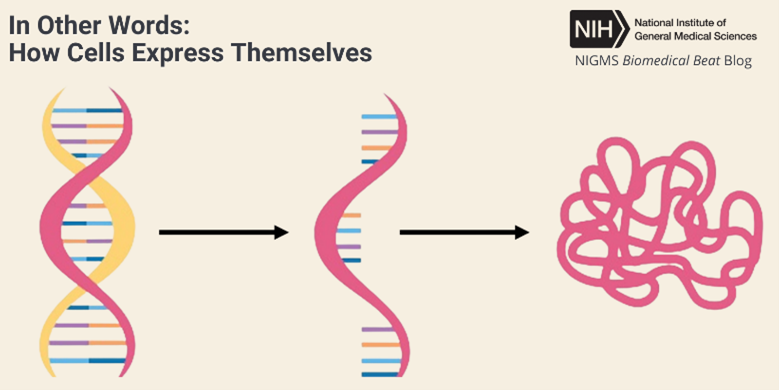
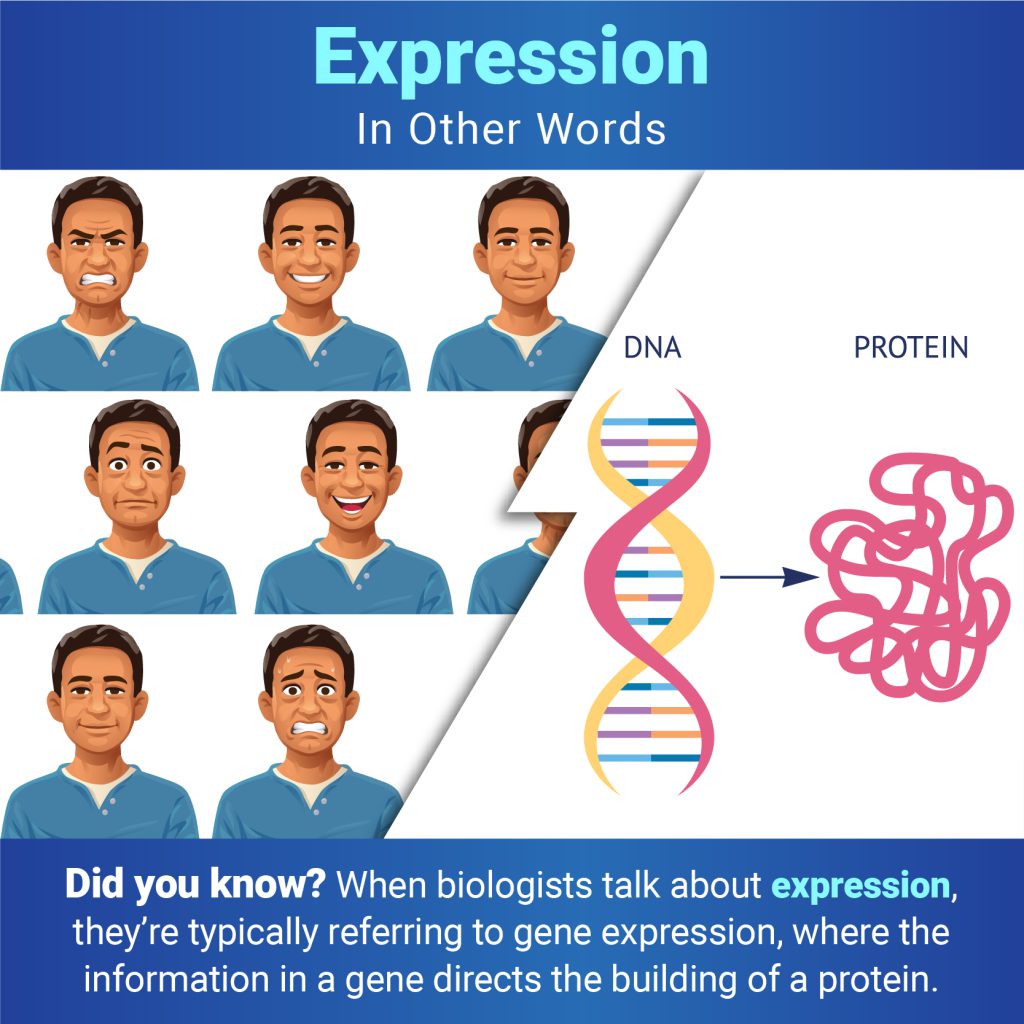
Continue reading “Hunting Disease-Causing Genetic Variants”In Other Words: How Cells Express Themselves
When you encounter the word expression, you may think of a smile, a grimace, or another look on someone’s face. But when biologists talk about expression, they typically mean the process of gene expression—when the information in a gene directs protein synthesis. Proteins are essential for virtually every process in the human body.
Public Alerted to Omicron in New Mexico Through Quick Detection
Over the past 2 years, you’ve probably heard a lot about the spread of SARS-CoV-2—the virus that causes COVID-19—and the emergence of variants. The discovery and tracking of these variants is possible thanks to genomic surveillance, a technique that involves sequencing and analyzing the genomes of SARS-CoV-2 virus particles from many COVID-19 patients. Genomic surveillance has not only shed light on how SARS-CoV-2 has evolved and spread, but it has also helped public health officials decide when to introduce measures to help protect people.
In December 2021, the NIGMS-supported SARS-CoV-2 genomic surveillance program at the University of New Mexico Health Science Center (UNM HSC) in Albuquerque detected the first known case of the Omicron variant in the state, which enabled a rapid public health response. The program’s co-leaders, assistant professors Darrell Dinwiddie, Ph.D., and Daryl Domman, Ph.D., were watching on high alert for it to enter New Mexico, and when it did, they were poised to quickly identify it:
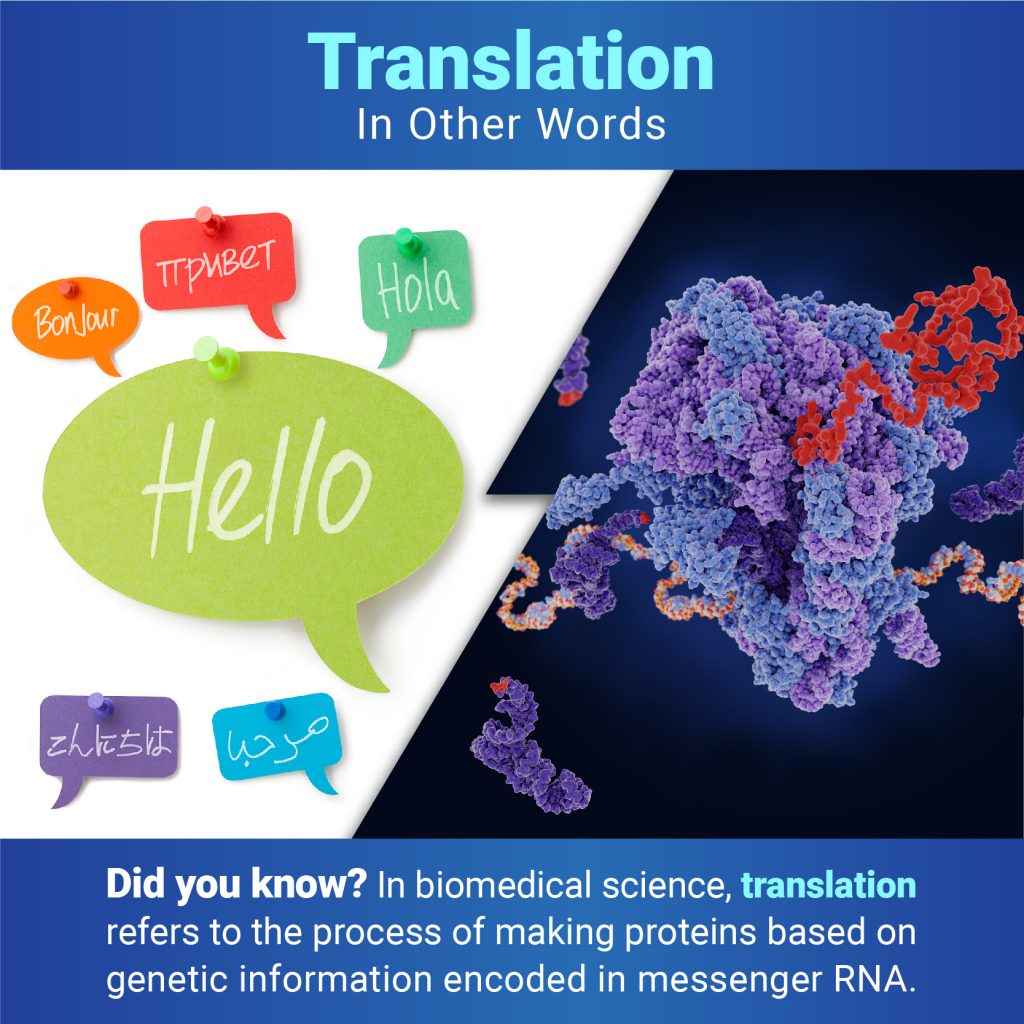
Continue reading “Public Alerted to Omicron in New Mexico Through Quick Detection”In Other Words: Translation Isn’t Only for Languages
In everyday use, most people understand translation to mean converting words from one language to another. But when biologists talk about translation, they mean the process of making proteins based on the genetic information encoded in messenger RNA (mRNA). Proteins are essential for virtually every process in our bodies, from transporting oxygen to defending against infection, so translation is vital for keeping us alive and healthy.
Quiz: Are You a Genetics Genius?
Genes are segments of DNA. They contain instructions for building one or more molecules that help the body work. Researchers in the field of genetics study genes and heredity—how certain traits are passed from parents to their offspring through DNA. NIGMS supports many scientists who investigate the genetics of people and research organisms to better understand human health and disease.
Take our quiz below to test how much you know about genetics. For more quizzes and other fun learning tools, visit our activities and multimedia webpage.
Continue reading “Quiz: Are You a Genetics Genius?”