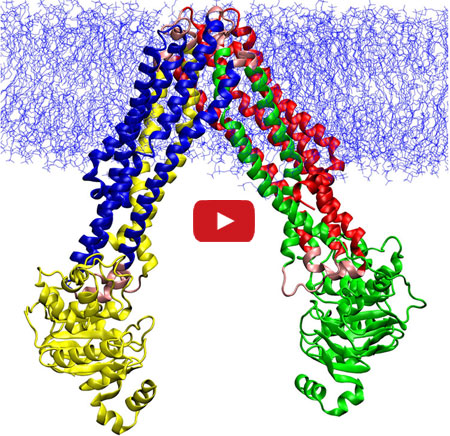
The discovery of a compound that slows the natural degradation of insulin in mice opens up a new area of investigation in the search for drugs to treat diabetes. The research team, which included David Liu ![]() and Alan Saghatelian of Harvard University, Markus Seeliger of Stony Brook University School of Medicine, and Wei-Jen Tang
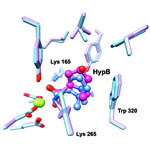
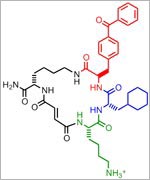
and Alan Saghatelian of Harvard University, Markus Seeliger of Stony Brook University School of Medicine, and Wei-Jen Tang ![]() of the University of Chicago focused on insulin-degrading enzyme, or IDE. Using a method called DNA-templated synthesis, the scientists made 14,000 small molecules and found one that bound to the enzyme, suggesting it might modulate the enzyme’s activity. Work in test tubes and in animal models confirmed this—and showed that blocking IDE activity improved insulin levels and glucose tolerance. The researchers also learned that the enzyme is misnamed: In addition to insulin, it degrades two other hormones involved in glucose regulation.
of the University of Chicago focused on insulin-degrading enzyme, or IDE. Using a method called DNA-templated synthesis, the scientists made 14,000 small molecules and found one that bound to the enzyme, suggesting it might modulate the enzyme’s activity. Work in test tubes and in animal models confirmed this—and showed that blocking IDE activity improved insulin levels and glucose tolerance. The researchers also learned that the enzyme is misnamed: In addition to insulin, it degrades two other hormones involved in glucose regulation.
NIGMS’ Peter Preusch says, “This is a very interesting fusion of chemical methods and biology that has uncovered new basic science findings about insulin processing with potential clinical impact.”
This work also was funded by NIH’s National Cancer Institute and the Office of the Director.
Learn more:
Harvard University News Article ![]()
Chemistry of Health Booklet



 Exposure to hypochlorous acid causes bacterial proteins to unfold and stick to one another, leading to cell death. Credit: Video segment courtesy of the American Chemistry Council. View video
Exposure to hypochlorous acid causes bacterial proteins to unfold and stick to one another, leading to cell death. Credit: Video segment courtesy of the American Chemistry Council. View video