Have you ever noticed plastic utensils or paper plates labeled as “biodegradable” and wondered what that meant? Materials are biodegradable when microorganisms such as bacteria can break them down into their building blocks.
Continue reading “Science Snippet: Breaking Down Biodegradability”Category: Chemistry, Biochemistry and Pharmacology
Martin Burke: Replacing Lost Proteins to Treat Disease
As a medical student, Martin Burke, M.D., Ph.D., helped care for a young college student with cystic fibrosis (CF), an inherited disease that affects the body’s ability to make sweat and mucus. Dr. Burke had just studied CF in class, so he relayed what he had learned to her. He had a lot of information to give—doctors and researchers know the exact amino acid changes in an ion channel protein called cystic fibrosis transmembrane conductance regulator (CFTR) that cause CF.
“At one point in the conversation, she stopped me and said, ‘It sounds like you know exactly what’s wrong with me, so why can’t you fix it?’” Dr. Burke, now the May and Ving Lee Professor for Chemical Innovation at University of Illinois Urbana-Champaign (UIUC), never forgot this question. In fact, it’s inspired his career-long search for new ways to develop therapies for diseases without effective treatment options.
Continue reading “Martin Burke: Replacing Lost Proteins to Treat Disease”In Other Words: What Being Unionized Means for Molecules
Did you know that molecules can be unionized? But it doesn’t mean they form a labor union. In chemistry, unionized (pronounced “un-ionized”) is the opposite of ionized, which means “electrically charged.”

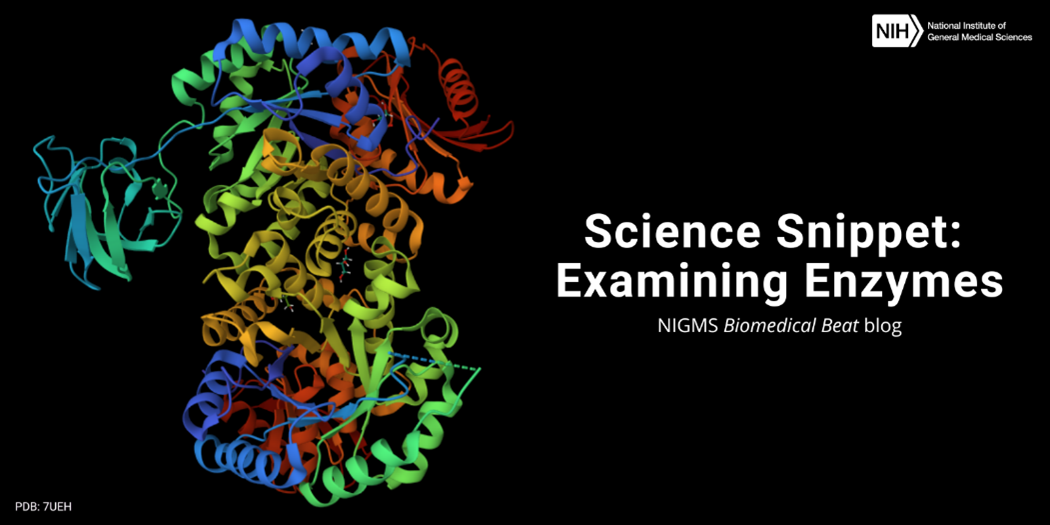
Science Snippet: Examining Enzymes
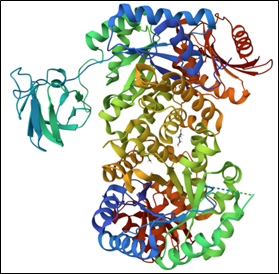
Every day, our cells must produce all the various molecules they need to stay alive. But the chemical reactions to create these molecules can’t occur without help—which is where enzymes come in. Enzymes are biological catalysts, meaning they speed up the rate of specific chemical reactions by reducing the amount of energy needed for the reaction to occur. Most enzymes are proteins, but some RNA molecules can also act as enzymes.
Thousands of different enzymes catalyze the vast range of reactions that take place within cells, but each enzyme typically supports one of the following types of tasks:
Continue reading “Science Snippet: Examining Enzymes”Amie Fornah Sankoh Achieves a Scientific Dream

“I wanted to give up so many times. Although I tried to remain positive, I never thought I’d be able to finish my Ph.D. But I made it, and I’m extremely proud of myself,” says Amie Fornah Sankoh, Ph.D., a research scientist with Dow Chemical Company who received NIGMS support as a graduate student.
Human and Plant Communication
Dr. Sankoh has loved science and mathematics since she was just a child growing up in Sierra Leone. When she was 3 years old, Dr. Sankoh became deaf from a childhood disease. Math, unlike other subjects, is very visual, which played a part in her interest in it. “Before I learned American Sign Language when I was 15 years old, I could only understand one language: mathematics,” Dr. Sankoh says.
Continue reading “Amie Fornah Sankoh Achieves a Scientific Dream”Developing Low-Cost Lab Techniques: Q&A With Abraham Badu-Tawiah

“I never thought I could make an impact on chemistry and students’ lives. But now, I’m the head of a lab with several Ph.D. and undergraduate students and a postdoctoral researcher; and we’re developing simple, low-cost lab techniques that can be adopted by labs across the world,” says Abraham Badu-Tawiah, Ph.D., the Robert K. Fox Professor of Chemistry at Ohio State University in Columbus. We talked with Dr. Badu-Tawiah about his career progression, research, and advice for students hoping to launch a career in science.
Q: How did you get started on the path to a career in science?
A: In Ghana, where I grew up, education works differently than in the United States. High school students are assigned subjects to study primarily based on their grades, and once assigned a subject, it’s difficult to switch. I was assigned to math, physics, and chemistry, which put me on a path toward being an engineer. I was happy to be studying science, but after the death of my brother, I wanted to study medicine more than engineering.
Continue reading “Developing Low-Cost Lab Techniques: Q&A With Abraham Badu-Tawiah”What Do Fats Do in the Body?
It’s common knowledge that too much cholesterol and other fats can lead to disease and that a healthy diet involves watching how much fatty food we eat. However, our bodies need a certain amount of fat to function—and we can’t make it from scratch.
Triglycerides, cholesterol, and other essential fatty acids—the fats our bodies can’t make on their own—store energy, insulate us, and protect our vital organs. They act as messengers, helping proteins do their jobs. They also start chemical reactions that help control growth, immune function, reproduction, and other aspects of basic metabolism. Fats also help the body stockpile certain nutrients. Vitamins A, D, E, and K, for example, are stored in the liver and in fatty tissues.
The cycle of making, breaking, storing, and using fats is at the core of how all animals, including humans, regulate their energy. An imbalance in any step can result in disease. For instance, having too many triglycerides in our bloodstream raises our risk of clogged arteries, which can lead to heart attack and stroke.
Continue reading “What Do Fats Do in the Body?”Inspiring the Next Generation of Scientists Through CityLab

“Many of the students we work with don’t have access to a laboratory through their local schools. For them, CityLab is their first exposure to a laboratory environment—these are hugely important moments for these kids,” says Carl Franzblau, Ph.D., the founder of CityLab at Boston University (BU). CityLab was established more than 30 years ago as a science education outreach program for precollege students and teachers through a partnership between the Chobanian & Avedisian School of Medicine and the Wheelock College of Education & Human Development at BU.
“Since our first Science Education Partnership Award (SEPA) grant in 1991, our mission has been to inspire students to consider careers in the biomedical sciences and broaden the opportunities that are available to them,” says Carla Romney, D.Sc., the director of research for CityLab. Continuous SEPA funding since 1991 has allowed CityLab to fulfill its mission and provide students with state-of-the-art biotechnology laboratory facilities and curricula.
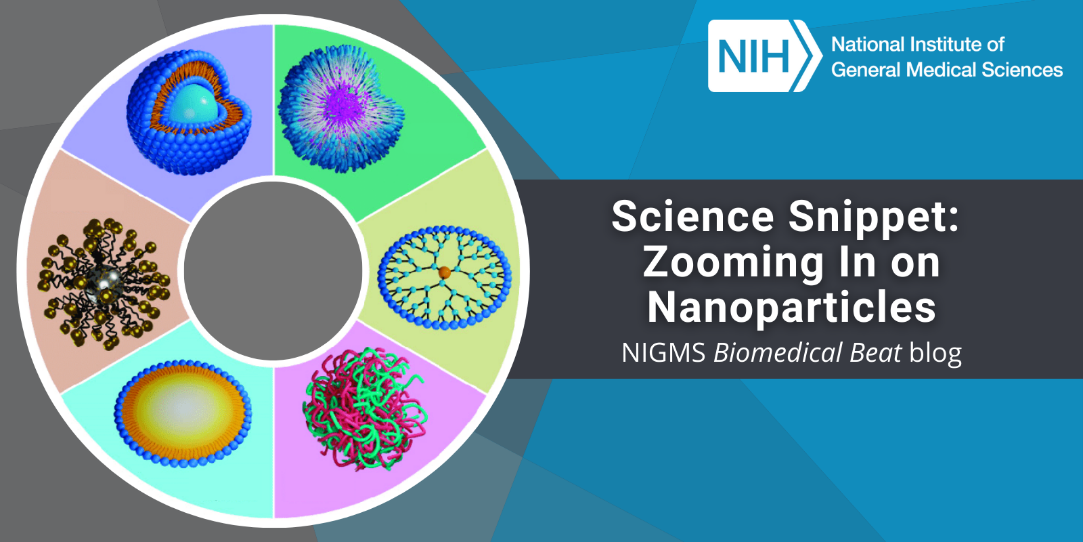
Continue reading “Inspiring the Next Generation of Scientists Through CityLab”Science Snippet: Zooming In on Nanoparticles
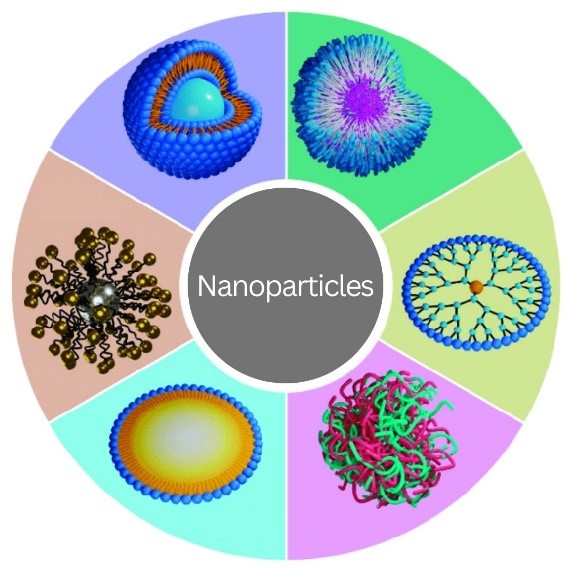
Nanoparticles may sound like gadgets from a science fiction movie, but they exist in real life. They’re particles of any material that are less than 100 nanometers (one-billionth of a meter) in all dimensions. Nanoparticles appear in nature, and humans have, mostly unknowingly, used them since ancient times. For example, hair dyeing in ancient Egypt involved lead sulfite nanoparticles, and artisans in the Middle Ages added gold and silver nanoparticles to stained-glass windows. Over the past several decades, researchers have studied nanoparticles for their potential uses in many fields, from computer engineering to biology.
A nanoparticle’s properties can differ significantly from those of larger pieces of the same material. Properties that may change include:
Continue reading “Science Snippet: Zooming In on Nanoparticles”Making Microprotein Discoveries With Alan Saghatelian

“There aren’t many professions that can provide this much opportunity for learning, especially when it comes to understanding how our bodies work. I really love what I do—I wouldn’t trade it for anything,” says Alan Saghatelian, Ph.D., a professor in the Clayton Foundation Laboratories for Peptide Biology at the Salk Institute for Biological Studies in La Jolla, California. From studying new facts and experimental techniques to adopting new ways of thinking, researchers never stop learning, and Dr. Saghatelian credits his love for learning and exploring as reasons why he’s perfectly suited for science. He’s used these passions to build a successful career in biochemistry.
From Chemistry to Biology
Dr. Saghatelian’s love for chemistry began when he was young. He was drawn to how predictable it could be: Mix two chemical compounds in the same way and they’ll always combine to form the same substance, as dictated by the rules of chemistry.
Continue reading “Making Microprotein Discoveries With Alan Saghatelian”