Do you like to find new uses for old things? Like weaving old shirts into a rug, repurposing bottles into candle holders or turning packing crates into tables? Katie Gostic, a University of California, Los Angeles (UCLA) graduate student, likes finding new uses for old data. She channeled this interest when she analyzed existing data to study whether childhood exposure to flu affects a person’s future immunity to the disease.

As an undergraduate student at Princeton University, Gostic was originally pursuing a degree in engineering. Her focus shifted to biology after taking an infectious disease modeling class. Gostic’s background in math and programming allows her to take large, complex pre-existing data sets and reanalyze them using new tools and methods. The result: Information that wasn’t accessible when the data were first collected.
Now a graduate researcher in the ecology and evolutionary biology lab of James Lloyd-Smith ![]() , Gostic studies infectious diseases. The lab builds mathematical models to investigate zoonotic diseases—diseases that animals can transmit to humans but that humans don’t frequently spread between each other. Examples include diseases caused by Leptospira, a type of bacteria that infects household pets and many other animals, and monkeypox, a virus whose transmission to humans is increasing since the eradication of smallpox. The lab also studies bird flus, a category of flu viruses that infect birds and other animals and only occasionally jump to people. A very small number of cases of human-to-human transmission of bird flus have been recorded. However, if a bird flu virus mutated in a way that allowed it to spread among humans, it could cause a pandemic.
, Gostic studies infectious diseases. The lab builds mathematical models to investigate zoonotic diseases—diseases that animals can transmit to humans but that humans don’t frequently spread between each other. Examples include diseases caused by Leptospira, a type of bacteria that infects household pets and many other animals, and monkeypox, a virus whose transmission to humans is increasing since the eradication of smallpox. The lab also studies bird flus, a category of flu viruses that infect birds and other animals and only occasionally jump to people. A very small number of cases of human-to-human transmission of bird flus have been recorded. However, if a bird flu virus mutated in a way that allowed it to spread among humans, it could cause a pandemic.
Gostic became involved in a project studying bird flus through a bit of an “accident,” she says. It began when she heard Michael Worobey of the University of Arizona, Tucson, give a seminar at UCLA where he introduced the idea that childhood exposure to seasonal flu strains could potentially provide immunity against future exposure to certain bird flus or other animal-origin flu viruses. In the past, researchers thought that humans wouldn’t have any immunity against flu viruses that came from animals. But Worobey’s hypothesis could explain why certain demographic age groups have been less susceptible to particular animal-origin strains, such as the 1918 Spanish flu pandemic or H5N1 and H7N9 (two bird flus with pandemic potential). “I was really excited after the talk,” recalls Gostic. “I was like, ‘Wow this could be a really big deal!’”
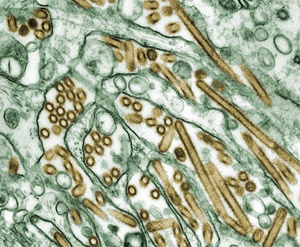
After the seminar, Gostic and Lloyd-Smith discussed the idea of collaborating with Worobey to work on a follow-up project to study if people’s childhood exposure to seasonal flu affects their immune system’s ability to fight off bird flus later in life. Worobey, now Gostic’s close mentor, agreed to collaborate. So with the help of labmate Monique Ambrose, the research team collected data from the World Health Organization (WHO), the Hong Kong Centre for Health Protection, FluTrackers and other sources. Much of the data they used is routinely collected by governments and agencies working with WHO.
The group’s study, published in Science and featured on the NIH Director’s Blog, reconstructed an almost 100-year history of seasonal flu in six countries and then compiled that with data on all known human cases of H5N1 and H7N9 that also reported the infected people’s ages. The researchers found the type of hemagglutinin protein (the “H” in H5N1 and H7N9) on the seasonal flu strain a person had likely first encountered during childhood influenced whether that person would be affected by a particular bird flu strain later in life. This finding is important for understanding who may have more protection against future pandemic flu strains and for creating new vaccines.
Gostic says she enjoys this kind of research because it uses her math and computer skills and has real-world impact. She also likes sharing her knowledge and skills with others. For example, she consults informally with her fellow graduate students to help them with coding problems and is a teaching assistant for several undergraduate classes. She has helped develop a curriculum about evolution designed for girls in elementary and middle school.
Her advice to others looking to do biology research is, “Take math classes and statistics classes, and learn to code. Look for opportunities or mentors who are going to help you use those skills to analyze biological data.” Gostic’s career goal: Become a faculty member at an institution where she can stay in research and continue teaching.
Gostic is funded by an NIGMS T32 predoctoral training grant that prepares graduate students for research or research-related careers in a variety of basic biomedical science areas.
More Biomedical Beat Stories




Thank you so much for writing an article that I as a layperson can understand. I am an intelligent, college educated woman but do not “speak Science”. I appreciate an article that explains something adequately without getting too bogged down in science-y details, yet is not condescending in tone. Great job on a topic I didn’t know could be so interesting!!