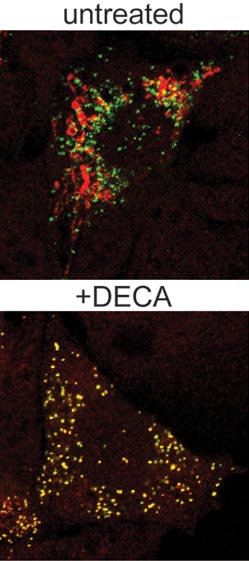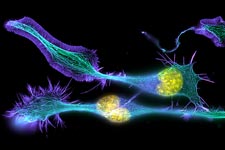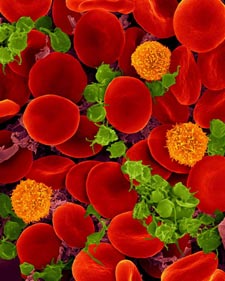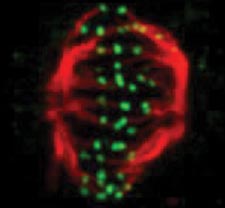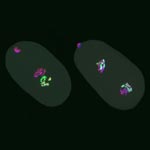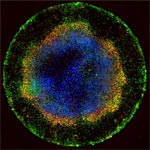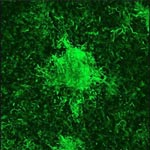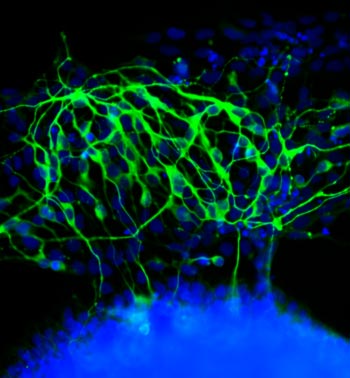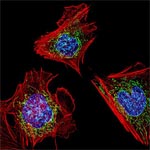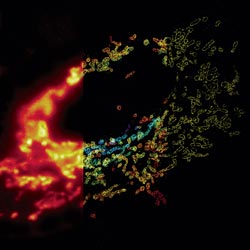
Much as a photographer brings distant objects into focus with a telephoto lens, scientists can now see previously indistinct cellular components as small as a few billionths of a meter (nanometers). By overcoming some of the limitations of conventional optical microscopy, a set of techniques known as super-resolution fluorescence microscopy has changed once-blurry images of the nanoworld into well-resolved portraits of cellular architecture, with details never seen before in biology. Reflecting its importance, super-resolution microscopy was recognized with the 2014 Nobel Prize in chemistry.
Using the new techniques, scientists can observe processes in living cells across space and time and study the movements, interactions and roles of individual molecules. For instance, they can identify and track the proteins that allow a virus to invade a cell or those that enable tumor cells to migrate to distant parts of the body in metastatic cancer. The ability to analyze individual molecules, rather than collections of molecules, allows scientists to answer longstanding questions about cellular mechanisms and behavior, such as how cells move along a surface or how certain proteins interact with DNA to regulate gene activity. Continue reading “Field Focus: Bringing Biology Into Sharper View with New Microscopy Techniques”