
The power of computer code has been a longtime fascination for Tomas Helikar, Ph.D., a professor of biochemistry at the University of Nebraska-Lincoln (UNL). In college, when he learned he could use that power to help researchers better understand biology and improve human health, Dr. Helikar knew he’d found his ideal career. Since then, he’s built a successful team of scientists studying the ways we can use mathematical models in biomedical research, such as creating a digital replica of the immune system that could predict how a patient will react to infectious microorganisms and other pathogenic insults.
A Career in Computational Biology
Dr. Helikar first became involved in computer science by learning how to build a website as a high school student. He was amazed to learn that simple lines of computer code could be converted into a functional website, and he felt empowered knowing that he had created a real product from his computer.
It wasn’t surprising, then, that Dr. Helikar entered the University of Nebraska Omaha as an undergraduate to study computer science and computer engineering. In his sophomore year, while considering job prospects as a computer scientist, his professor Jim Rogers, Ph.D., sought undergraduate students to help build a mathematical model of biochemical pathways. Although Dr. Helikar was unsure what that entailed, he took the position. The decision to join Dr. Rogers’ lab entirely changed his career trajectory.
“That project opened my eyes to this world at the intersection of biology, mathematics, and computer science, and I suddenly saw the career opportunities,” Dr. Helikar explains. He realized that he could apply his passion for developing software in a way that could make a difference for human health. “I found myself excited again by the prospect of creating meaningful products with just computer code,” he says. With that, he changed his major and ultimately graduated with a bachelor’s degree in bioinformatics.
The idea of graduate school initially intimidated Dr. Helikar, but, with Dr. Rogers’ encouragement, he decided to pursue his Ph.D. in the Rogers lab at the University of Nebraska Medical Center (UNMC) in Omaha. He continued the computational biology project he helped with as an undergraduate, studying how cells process and respond to the signals they constantly receive from their environment. Specifically, he tested the hypothesis that connected pathways, called networks, are important for information processing and decision-making in cells.

As a part of this project, Dr. Helikar and the rest of the team built a mathematical model of the networks using published information on the proteins involved. Then, they ran simulations of the model, exposing virtual cells to millions of combinations of signals to learn how they respond. One finding from this study was that cells respond to these millions of signal combinations with only a few potential outcomes, such as cell division, death, or differentiation—the process by which an unspecialized cell becomes a specialized cell with a specific function.
Dr. Helikar also began exploring the possibility of applying models of biochemical networks to disease-specific research. In collaboration with Hamid Band, M.D., Ph.D., he built a model of the signaling networks involved in breast cancer to identify proteins that could be potential therapeutic targets. As with the first model, he ran simulations to learn if altering the function or activity of a specific protein within the networks yielded the desired outcome, which in this case was cell death.
Given his success and ongoing projects, Dr. Helikar stayed at UNMC as a postdoctoral researcher with Dr. Rogers after completing his Ph.D. During this time, he continued building new models and developed an interest in human immunology.
Your Digital Twin
After moving to his current position at UNL, Dr. Helikar explored how he could apply his strengths in model development to his new interest in immunology. He focused his attention on a specific type of white blood cell called the helper T cell. Researchers have learned that there are several types of helper T cells, and each has a distinct job. For example, one type might be specialized for fighting viruses, while another is equipped to target bacteria. Unspecialized helper T cells have complex signaling networks that help them decide which type to become. Dr. Helikar’s lab is using mathematical models to understand how different combinations of signals give rise to each type of helper T cell.
Researchers have also learned that differentiated helper T cells can be reprogrammed into one of the other types when exposed to different signal combinations. Dr. Helikar is interested in exploring if the entire immune system might be similarly malleable. If it is, doctors might one day be able to treat immunodeficiency disorders or autoimmune diseases, such as multiple sclerosis, with drug therapies that change how networks respond to signals.
Before researchers can create these types of treatments, though, they need a better understanding of the healthy immune system and a way to model it. Dr. Helikar and his lab are developing a digital twin of the immune system—a multiscale, multicellular model of a healthy immune system that can be tailored to represent an individual patient. It will include metabolic pathways, protein-level information, signal transduction networks within and among cells, and many types of patient-specific clinical data, such as infectious disease history, vaccine history, and number and types of white blood cells. With this information, the digital twin could predict how a specific person will respond to an infectious intruder, helping to identify people who are at high risk of infection or identify new or personalized treatments for immune-related conditions. Eventually, it could be used to understand how to adjust the immune responses in these patients to reduce risk.
Cell Collective
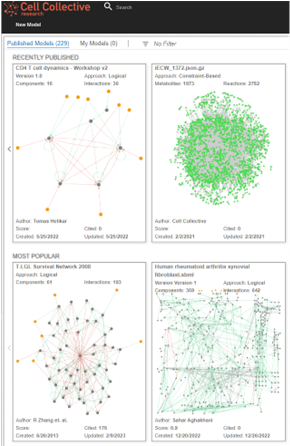
Creating simulations of biological systems, such as the digital twin, requires a lot of technical expertise, but Dr. Helikar wanted noncomputational biologists to be able to use and benefit from them. To tackle this goal, he developed an easy-to-use software called Cell Collective, which allows users ranging from experimental scientists and computational biologists to clinicians and even patients to build, visualize, and analyze simulations of biological systems.
To initially check that Cell Collective was easy enough to use, Dr. Helikar recruited undergraduate students in the biology department to test it. They were able to use the software without trouble and even learned about biochemistry in the process. Realizing the opportunity, Dr. Helikar developed and taught a new course on signal transduction pathways, using mathematical model building as the only teaching tool. “Students loved the course, and they were successfully learning the biology and the coding,” Dr. Helikar says.
Since then, Cell Collective has blossomed into more than just a research technology as hundreds of universities and high schools are using it as a teaching tool. The software has even been adapted into games for elementary school students as an early introduction to coding and modeling. “The most exciting part for me is that we’ve come full circle in the goal of accessible computational biology tools. Now, students from kindergarten through college are learning about science, modeling, and simulation through the same technology that scientists are using to make biomedical discoveries,” Dr. Helikar says.
Dr. Helikar’s work is funded by the NIGMS Maximizing Investigators’ Research Award program through grant R35GM119770.






Dr. Helikar’s transition from coding websites to modeling immune systems is impressive. It’s a reminder that we can always pivot in our careers to make a difference. Imagine if we could predict our immune system’s response during stressful work periods!