
The power of computer code has been a longtime fascination for Tomas Helikar, Ph.D., a professor of biochemistry at the University of Nebraska-Lincoln (UNL). In college, when he learned he could use that power to help researchers better understand biology and improve human health, Dr. Helikar knew he’d found his ideal career. Since then, he’s built a successful team of scientists studying the ways we can use mathematical models in biomedical research, such as creating a digital replica of the immune system that could predict how a patient will react to infectious microorganisms and other pathogenic insults.
A Career in Computational Biology
Dr. Helikar first became involved in computer science by learning how to build a website as a high school student. He was amazed to learn that simple lines of computer code could be converted into a functional website, and he felt empowered knowing that he had created a real product from his computer.
Continue reading “Building a Digital Immune System”




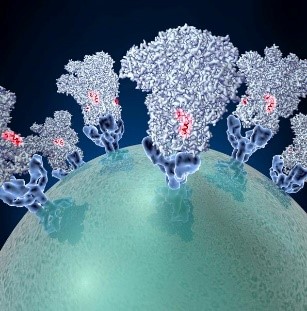 Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
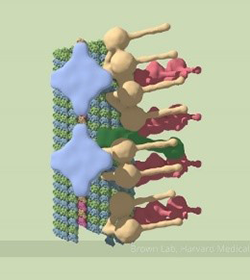 A partial model of a doublet microtubule. Credit: Veronica Falconieri.
A partial model of a doublet microtubule. Credit: Veronica Falconieri.
 A bioprint of the small air sac in the lungs with red blood cells moving through a vessel network supplying oxygen to living cells. Credit: Rice University.
A bioprint of the small air sac in the lungs with red blood cells moving through a vessel network supplying oxygen to living cells. Credit: Rice University.