Credit: New York Times article, Jan. 19, 2016.
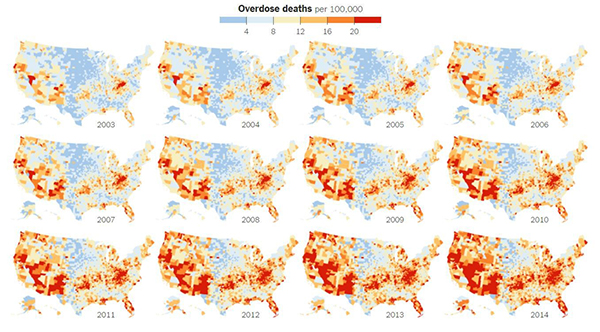
The United States is in the midst of an opioid overdose epidemic. The rates of opioid addiction, babies born addicted to opioids, and overdoses have skyrocketed in the past decade. No population has been hit harder than rural communities. Many of these communities are in states with historically low levels of funding from the National Institutes of Health (NIH). NIGMS’ Institutional Development Award (IDeA) program builds research capacities in these states by supporting basic, clinical, and translational research, as well as faculty development and infrastructure improvements. IDeA-funded programs in many states have begun prioritizing research focused on reducing the burden of opioid addiction. Below is a snapshot of three of these programs, and how they are working to help their communities:
Vermont Center on Behavior and Health
Because there are generally fewer treatment resources in rural areas compared to larger cities, it can take longer for people addicted to opioids in rural settings to get the care they need. The Vermont Center on Behavior and Health works to address this need and its major implications.
“One very disconcerting trend we’re seeing with this recent crisis is that opioid-addicted individuals are being placed on wait lists lasting months to a year without any kind of treatment,” says Vermont Center on Behavior and Health director Stephen Higgins. “And it’s very unlikely that anyone who is opioid addicted is just going to abstain while they are on a wait list.”
In urban areas, buprenorphine—an approved medication for opioid addiction that can prevent or reduce withdrawal symptoms—is generally dispensed by trained physicians at treatment clinics. Unfortunately, many rural communities don’t have enough physicians and clinics to serve patients in need. While waiting for treatment, patients are at risk of premature death, overdose, and contracting diseases such as HIV.
Stacey Sigmon, a faculty member in the Vermont Center on Behavior Health, has developed a method to help tackle this problem: a modified version of a tamper-proof device that delivers daily doses of buprenorphine. The advantage of using the modified device is that it makes each day’s dose available during a preprogrammed 3-hour window within the patient’s home, eliminating the need to visit a clinic.
During a study, participants in the treatment group received interim buprenorphine from the device. They also received daily calls to assess drug use, craving, and withdrawal. Participants in the control group didn’t receive buprenorphine. They remained on the waiting list of their local clinic and didn’t receive phone calls. The results, published in the New England Journal of Medicine (NEJM), indicate that the device works. Participants who received the interim buprenorphine treatment submitted a higher percentage of drug test specimens that were negative for opioids than those in the control group at 4 weeks (88 percent vs. 0 percent), 8 weeks (84 percent vs. 0 percent), and 12 weeks (68 percent vs. 0 percent). Sigmon and colleagues are currently testing the device with a much larger group of participants.
“This tool is now available to other rural states that are also being devastated by this crisis and are not so far along in beefing up treatment capacity,” says Higgins.
Continue reading “Americans Fighting the Opioid Crisis in Their Own Backyards”