If you’re looking for engaging ways to teach science from home, NIGMS offers a range of resources that can help.
 A SEPA-funded resource about microbes. Credit: University of Nebraska, Lincoln.
A SEPA-funded resource about microbes. Credit: University of Nebraska, Lincoln.
Our Science Education and Partnership Award (SEPA) webpage features free, easy-to-access STEM and informal science education projects for pre-K through grade 12. Aligned with state and national standards for STEM teaching and learning, the program has tools such as:
- Apps
- Interactives
- Online books
- Curricula and lesson plans
- Short movies
Students can learn about sleep, cells, growth, microbes, a healthy lifestyle, genetics, and many other subjects.
Continue reading “Explore Our Virtual Learning STEM Resources”

 Sohini Ramachandran, Brown University.
Sohini Ramachandran, Brown University. Hawaiian bobtail squid. Credit: Dr. Satoshi Shibata.
Hawaiian bobtail squid. Credit: Dr. Satoshi Shibata.
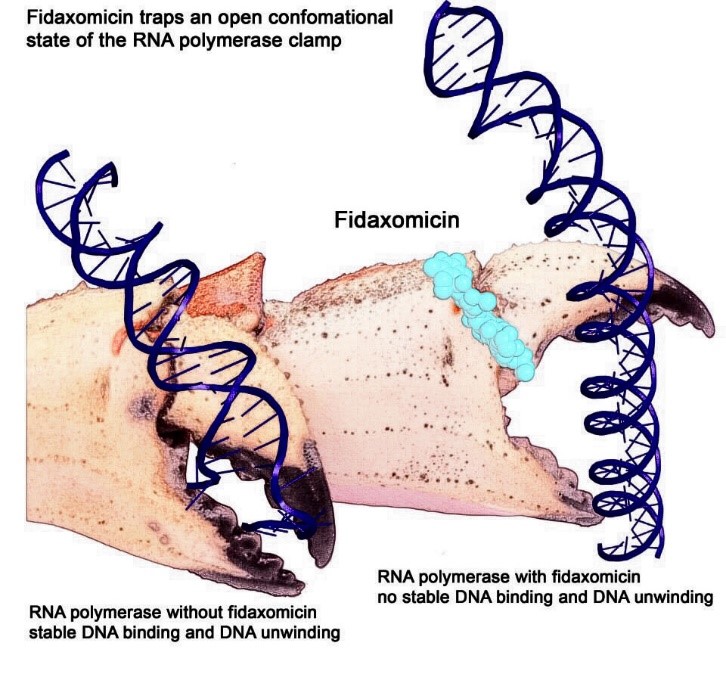 Artist interpretation of RNAP grasping and unwinding a DNA double helix. Credit: Wei Lin and Richard H. Ebright.
Artist interpretation of RNAP grasping and unwinding a DNA double helix. Credit: Wei Lin and Richard H. Ebright. Dr. Melissa Wilson.
Dr. Melissa Wilson.