
“I study the dance between a bacterium and its host. If we can decode the secrets of that dance—how the pathogen causes disease, and how the host fights back—we might be able to take advantage of vulnerabilities to improve our ability to combat infections,” says Víctor J. Torres, Ph.D., the C. V. Starr Professor of Microbiology at the New York University (NYU) Grossman School of Medicine in New York City.
Discovering and Pursuing a Passion for Science
Growing up, Dr. Torres never would have imagined his highly successful scientific career, especially since he didn’t have a strong interest in science. He entered the University of Puerto Rico, Mayagüez, in 1995, planning to participate in the Reserve Officers’ Training Corps and join the Air Force after graduation. He struggled during his first year of college and had to repeat several courses. In one of those courses, he met a fellow student who was planning to pursue a career in science—his now wife, Carmen A. Perez, M.D., Ph.D., who’s a radiation oncologist at NYU Langone. She shared with Dr. Torres some of the opportunities in science available to him, including the NIGMS-funded Maximizing Access to Research Careers (MARC) program at their university.
Continue reading “From MARC Student to MacArthur Fellow”



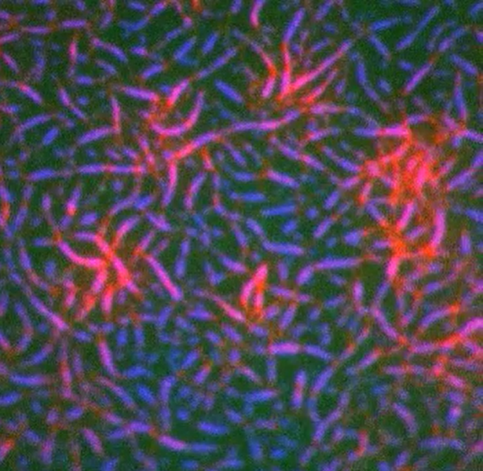

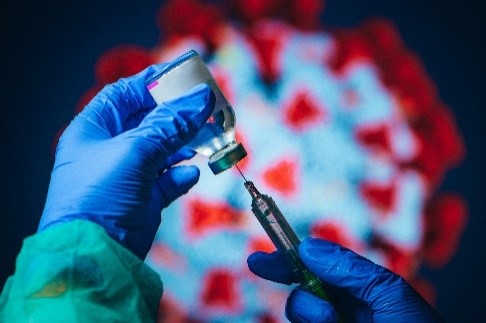 ACTIV clinical trials will evaluate the safety and efficacy of COVID-19 treatments and vaccines. Credit: iStock.
ACTIV clinical trials will evaluate the safety and efficacy of COVID-19 treatments and vaccines. Credit: iStock.
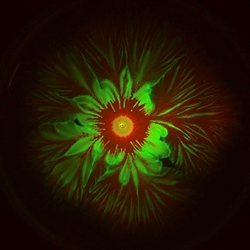 Credit: Liyang Xiong and Lev Tsimring, BioCircuits Institute, UCSD.
Credit: Liyang Xiong and Lev Tsimring, BioCircuits Institute, UCSD.
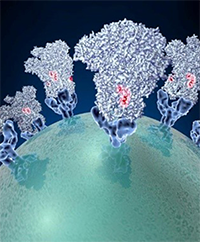 Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
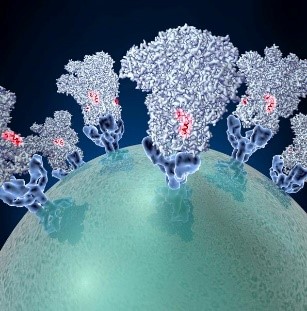 Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
Spike proteins on the surface of a coronavirus. Credit: David Veesler, University of Washington.
 Cover of Pathways student magazine.
Cover of Pathways student magazine.