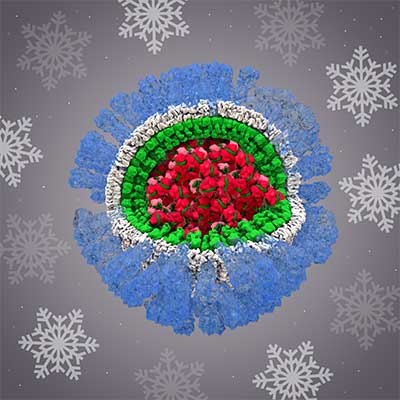
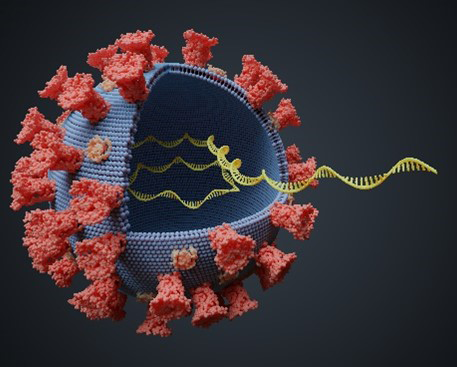
While she was in graduate school, Mandy Muller, Ph.D., became intrigued with viruses that are oncogenic, meaning they can cause cancer. At the time, she was researching human papillomaviruses (HPVs), which can lead to cervical and throat cancer, among other types. Now, as an assistant professor of microbiology at the University of Massachusetts (UMass) Amherst, Dr. Muller studies Kaposi sarcoma-associated herpesvirus (KSHV), which causes the rare AIDS-associated cancer Kaposi sarcoma.
A Continental Change
Dr. Muller has come a long way, both geographically and professionally, since her childhood in France. She was the first person in her family to graduate from high school, where she excelled in science, and went on to attend École Normale Supérieure (ENS) de Lyon, a research-oriented undergraduate institution in Lyon, France. “We spent weeks at a time in laboratory-based classes, working in real labs. That’s when I realized people could do research full-time, which caught my attention,” says Dr. Muller. She double-majored in biology and geology, and soon chose to focus her career on immunology and virology.
Continue reading “Investigating the Secrets of Cancer-Causing Viruses”





 Cover of Pathways student magazine.
Cover of Pathways student magazine.
 View the
View the